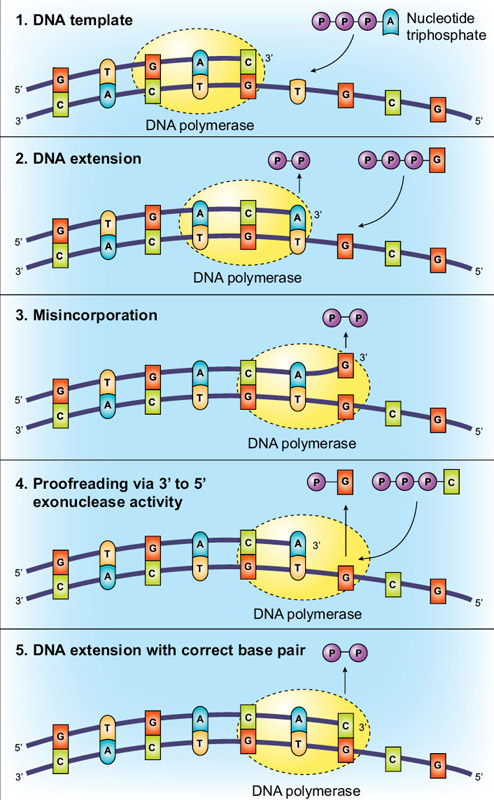
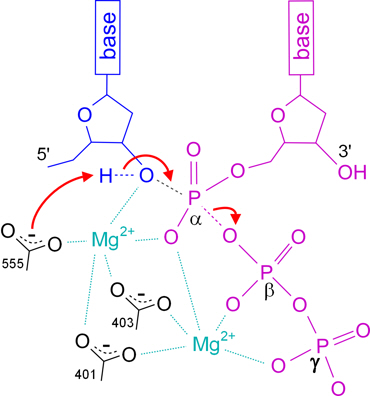
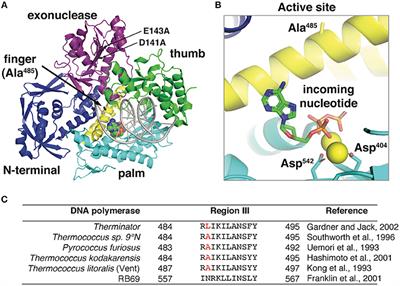
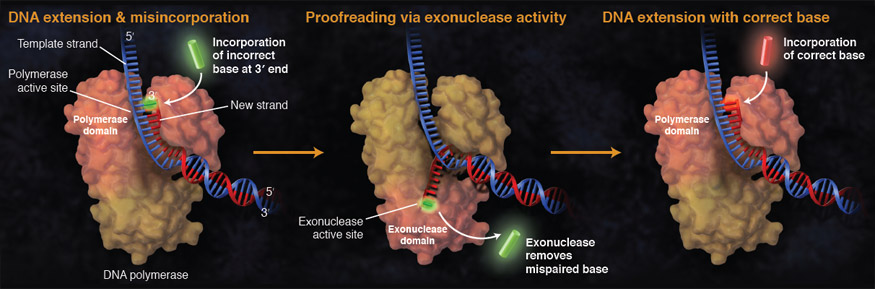
Figure 4 from Crystal Structure of the Catalytic α Subunit of E. coli Replicative DNA Polymerase III | Semantic Scholar
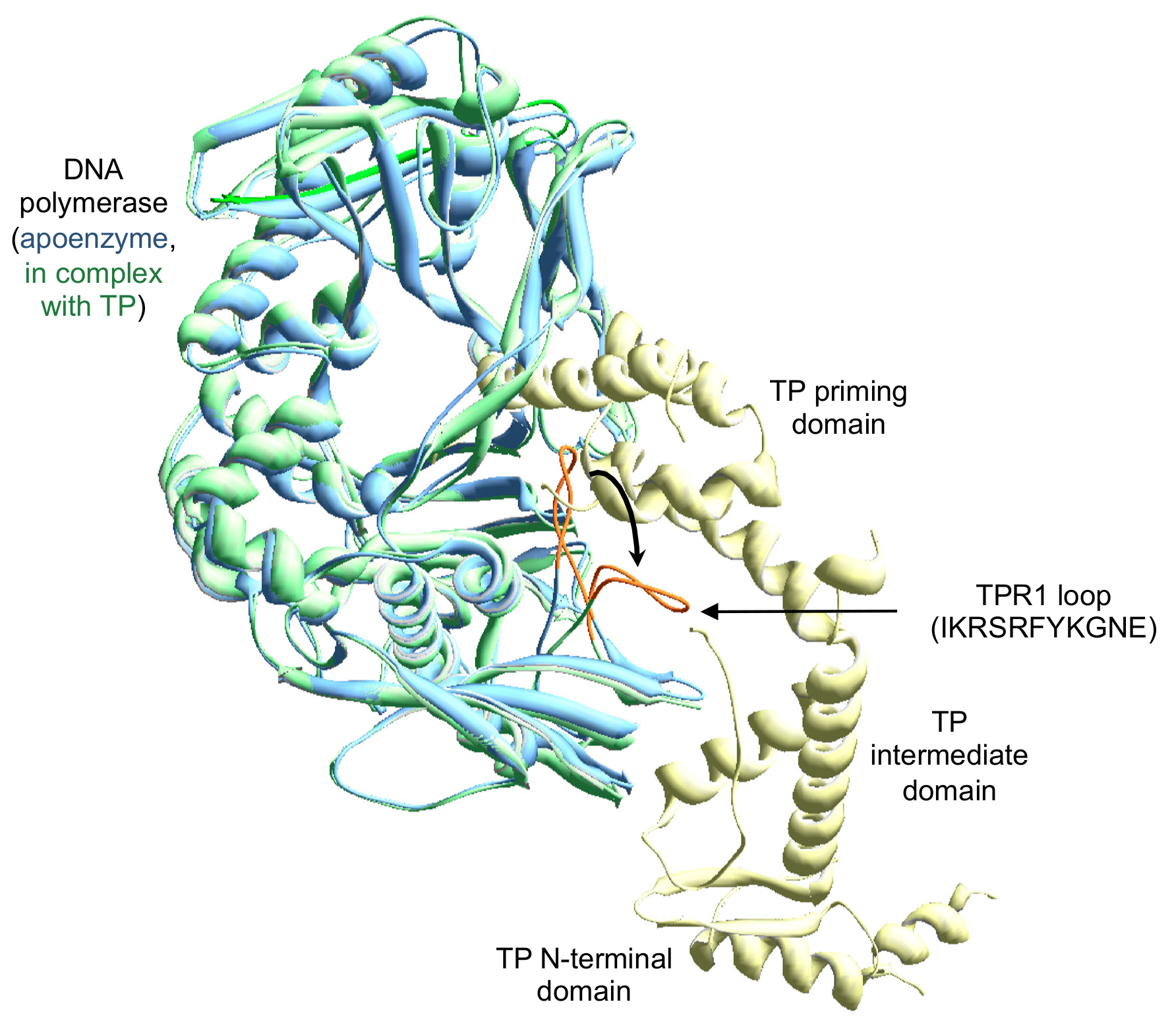
Biomolecules | Free Full-Text | The Loop of the TPR1 Subdomain of Phi29 DNA Polymerase Plays a Pivotal Role in Primer-Terminus Stabilization at the Polymerization Active Site

Insight into the mechanism of DNA synthesis by human terminal deoxynucleotidyltransferase | Life Science Alliance
A Model for Transition of 5′-Nuclease Domain of DNA Polymerase I from Inert to Active Modes | PLOS ONE

Critical Role of Magnesium Ions in DNA Polymerase β's Closing and Active Site Assembly | Journal of the American Chemical Society
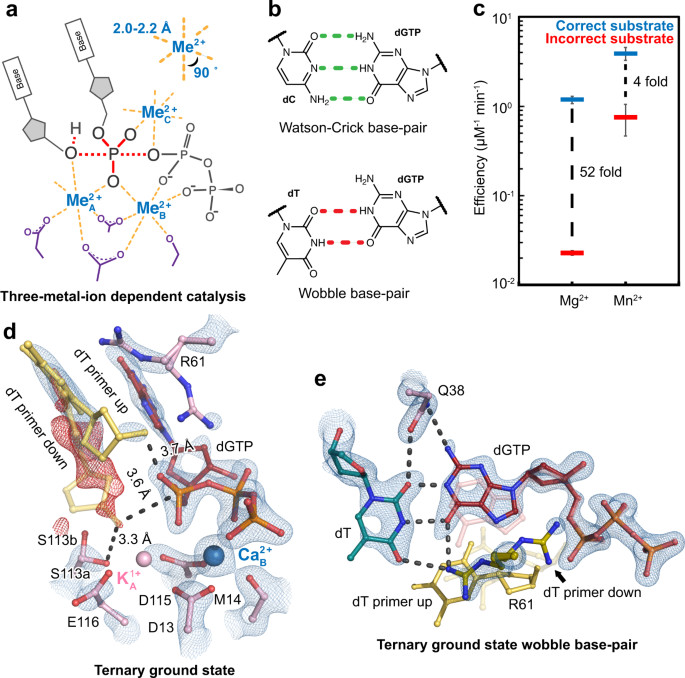
In crystallo observation of three metal ion promoted DNA polymerase misincorporation | Nature Communications

Critical Role of Magnesium Ions in DNA Polymerase β's Closing and Active Site Assembly | Journal of the American Chemical Society

The Structural Basis for Processing of Unnatural Base Pairs by DNA Polymerases - Marx - 2020 - Chemistry – A European Journal - Wiley Online Library